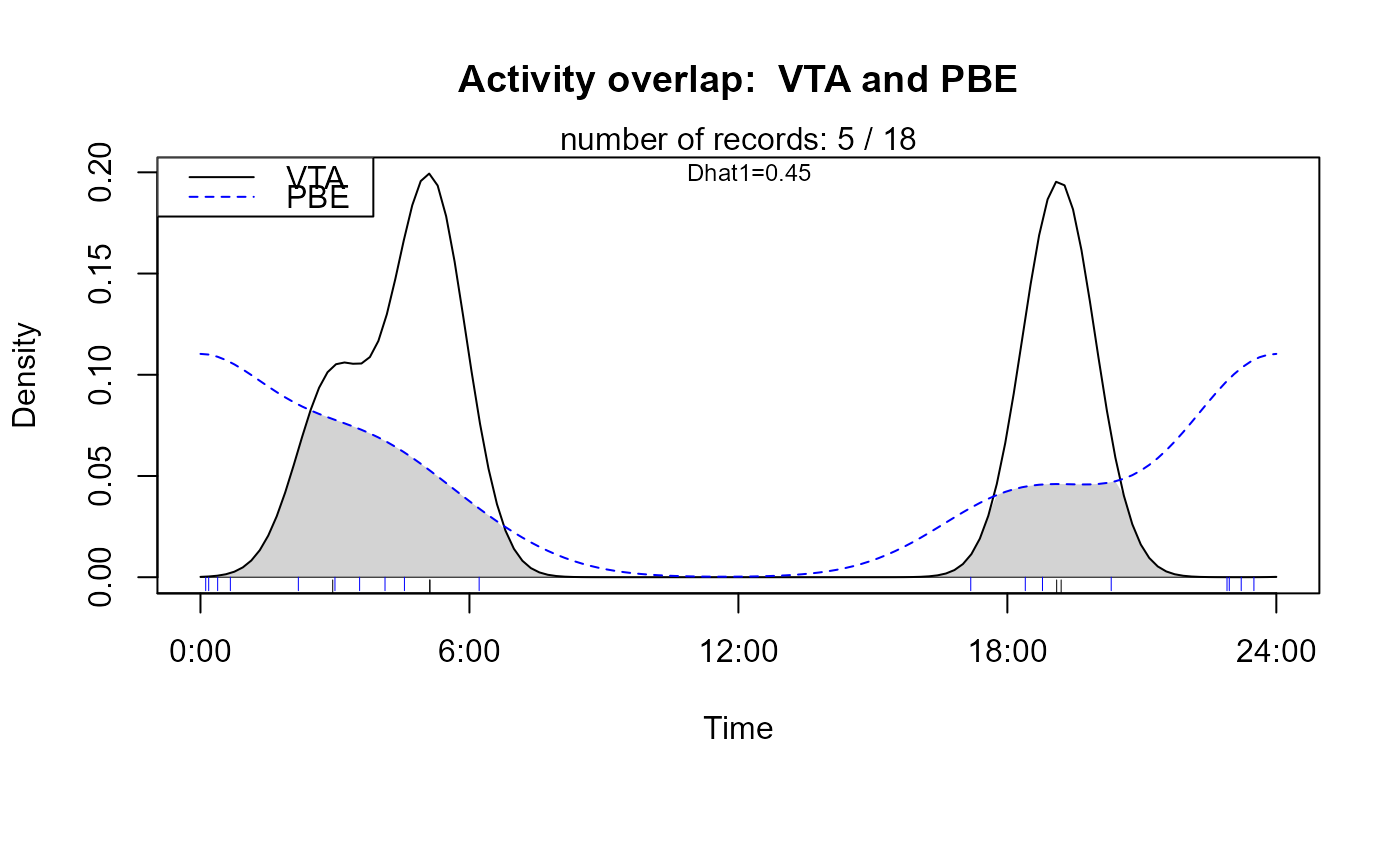
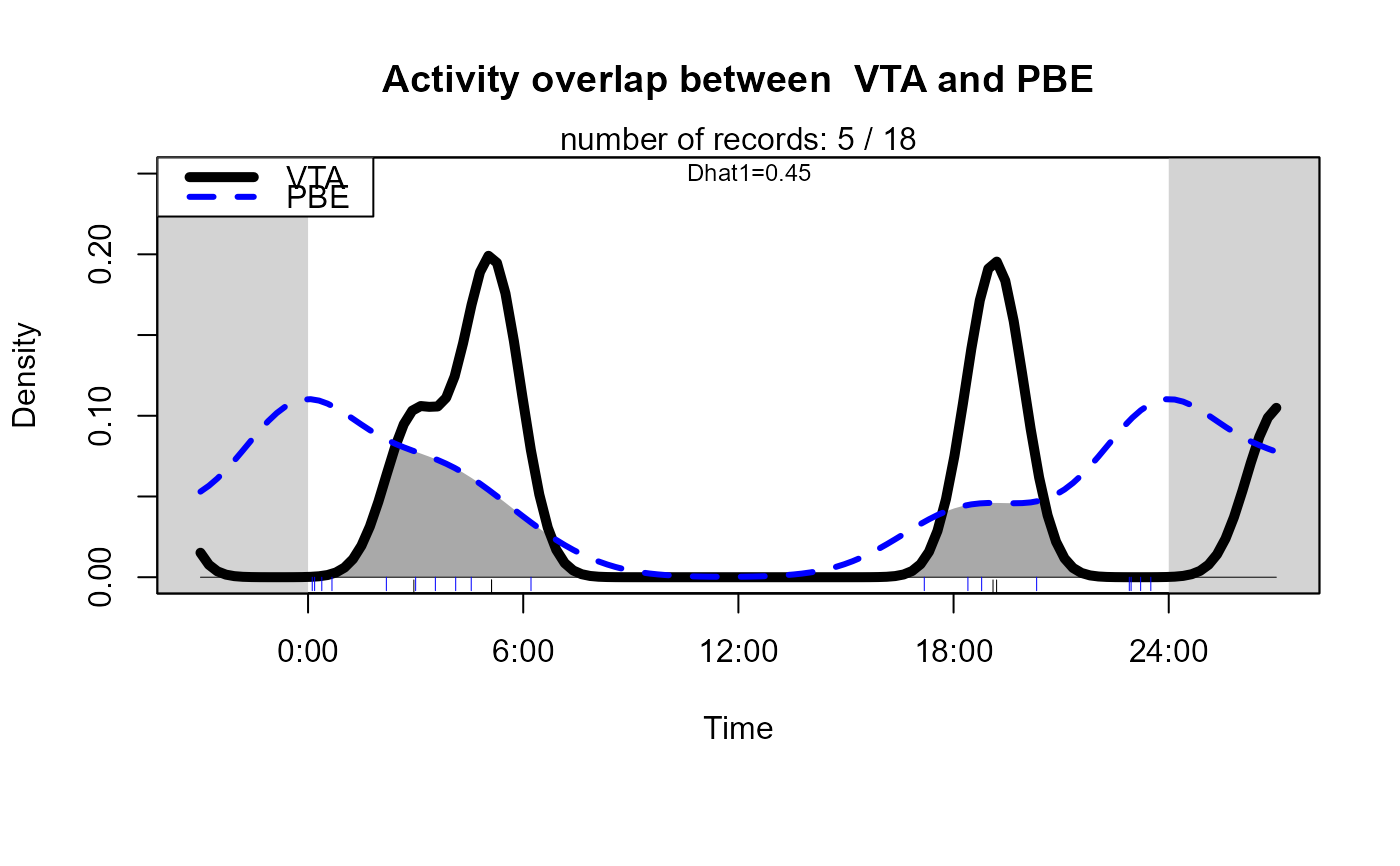
Plot overlapping kernel densities of two-species activities
Source:R/activityOverlap.R
activityOverlap.RdThis function plots kernel density estimates of two species' diel activity
data by calling the function overlapPlot from package
overlap. It further computes the overlap coefficient Dhat1 by calling
overlapEst.
activityOverlap(
recordTable,
speciesA,
speciesB,
speciesCol = "Species",
recordDateTimeCol = "DateTimeOriginal",
recordDateTimeFormat = "ymd HMS",
plotR = TRUE,
writePNG = FALSE,
addLegend = TRUE,
legendPosition = "topleft",
plotDirectory,
createDir = FALSE,
pngMaxPix = 1000,
add.rug = TRUE,
overlapEstimator = c("Dhat1", "Dhat4", "Dhat5"),
...
)Arguments
- recordTable
data.frame. the record table created by
recordTable- speciesA
Name of species 1 (as found in
speciesColof recordTable)- speciesB
Name of species 2 (as found in
speciesColof recordTable)- speciesCol
character. name of the column specifying species names in
recordTable- recordDateTimeCol
character. name of the column specifying date and time in
recordTable- recordDateTimeFormat
character. format of column
recordDateTimeColinrecordTable- plotR
logical. Show plots in R graphics device?
- writePNG
logical. Create pngs of the plots?
- addLegend
logical. Add a legend to the plots?
- legendPosition
character. Position of the legend (keyword)
- plotDirectory
character. Directory in which to create png plots if
writePNG = TRUE- createDir
logical. Create
plotDirectory?- pngMaxPix
integer. image size of png (pixels along x-axis)
- add.rug
logical. add a rug to the plot?
- overlapEstimator
character. Which overlap estimator to return (passed on to argument
typeinoverlapEst)- ...
additional arguments to be passed to function
overlapPlot
Value
Returns invisibly the data.frame with plot coordinates
returned by overlapPlot.
Details
... can be graphical parameters passed on to function
overlapPlot, e.g. linetype, linewidth,
linecol (see example below).
recordDateTimeFormat defaults to the "YYYY-MM-DD HH:MM:SS"
convention, e.g. "2014-09-30 22:59:59". recordDateTimeFormat can be
interpreted either by base-R via strptime or in
lubridate via parse_date_time (argument
"orders"). lubridate will be used if there are no "%" characters in
recordDateTimeFormat.
For "YYYY-MM-DD HH:MM:SS", recordDateTimeFormat would be either
"%Y-%m-%d %H:%M:%S" or "ymd HMS". For details on how to specify date
and time formats in R see strptime or
parse_date_time.
Note
Please be aware that the function (like the other activity... function of this package) use clock time, not solar time. If your survey was long enough to see changes in sunrise and sunset times, this may result in biased representations of species activity.
References
Mike Meredith and Martin Ridout (2018). overlap: Estimates of
coefficient of overlapping for animal activity patterns. R package version
0.3.2. https://CRAN.R-project.org/package=overlap
Ridout, M.S. and
Linkie, M. (2009) Estimating overlap of daily activity patterns from camera
trap data. Journal of Agricultural, Biological and Environmental Statistics,
14, 322-337.
Examples
if(requireNamespace("overlap")) {
# load record table
data(recordTableSample)
# define species of interest
speciesA_for_activity <- "VTA" # = Viverra tangalunga, Malay Civet
speciesB_for_activity <- "PBE" # = Prionailurus bengalensis, Leopard Cat
# create activity overlap plot (basic)
activityOverlap (recordTable = recordTableSample,
speciesA = "VTA", # = Viverra tangalunga, Malay Civet
speciesB = "PBE", # = Prionailurus bengalensis, Leopard Cat
writePNG = FALSE,
plotR = TRUE
)
# create activity overlap plot (prettier and with some overlapPlot arguments set)
activityOverlap (recordTable = recordTableSample,
speciesA = speciesA_for_activity,
speciesB = speciesB_for_activity,
writePNG = FALSE,
plotR = TRUE,
createDir = FALSE,
pngMaxPix = 1000,
linecol = c("black", "blue"),
linewidth = c(5,3),
linetype = c(1, 2),
olapcol = "darkgrey",
add.rug = TRUE,
extend = "lightgrey",
ylim = c(0, 0.25),
main = paste("Activity overlap between ",
speciesA_for_activity, "and",
speciesB_for_activity)
)
}